Macromolecular additive manufacturing
Making the proteins that living cells cannot make
I. Executive Summary
It might be possible to create multi-component proteins made from up to one hundred distinct parts, which would be over a ten-fold increase versus what can be made routinely with living cells, by devising a process called macromolecular additive manufacturing. This would entail:
- Multi-component proteins[1] that are built up piece by piece, where the first building block is attached to a solid material that functions as a ‘workbench’ so that additional pieces suspended in liquid can be flown across one at a time to grow the product.
- Complicated multi-component protein designs to be broken down into arrangements of standardized protein building blocks that snap together like Lego bricks.
These complex, multi-component proteins could enable artificial ‘antibodies’ that engage many biological targets simultaneously, vaccines made from multiple parts of infectious agents for robust immunity to many pathogens, or rapid prototyping of cell-free factories to synthesize chemicals with spatially organized enzymes.
Existing methods to make synthetic proteins are plagued with limitations at all levels of design and manufacturing processes:
- In silico design tools are not equipped to make multi-component proteins with many different parts.
- Ribosomal protein expression can only produce single proteins of a finite size.
- It is hard to know whether a new protein design will form successfully into the desired structure.
- It is difficult to add multiple new chemical functionalities because the proteins are free floating in liquid.
Consequently, the only large multi-component proteins with many different parts we can build are tweaks on evolved structures from nature.
Macromolecular additive manufacturing is fundamentally different from the existing self-assembling methods to design and make proteins because scaling from two components to hundreds would be straightforward, depending only on the number of sequential steps.
Creating this technology requires a coordinated effort with several research projects to build a cohesive macromolecular additive manufacturing system to demonstrate scalable synthesis of multi-component proteins with tens or even hundreds of parts. This program is split into three phases, will take roughly five years, and cost about five million dollars:
- Development of solid materials suitable for multi-component proteins and creation of possible protein building block designs (~21 months).
- Implementation of processes to build macromolecules on the solid material ‘workbench’ and testing of the protein building blocks (~21 months).
- Integration of the solid materials and building blocks into a continuous flow reactor that is user friendly to make multi-component proteins with up to one hundred parts (~18 months).
The roadmap below explains what this program is doing, the existing limitations, and why this is important in much more depth.
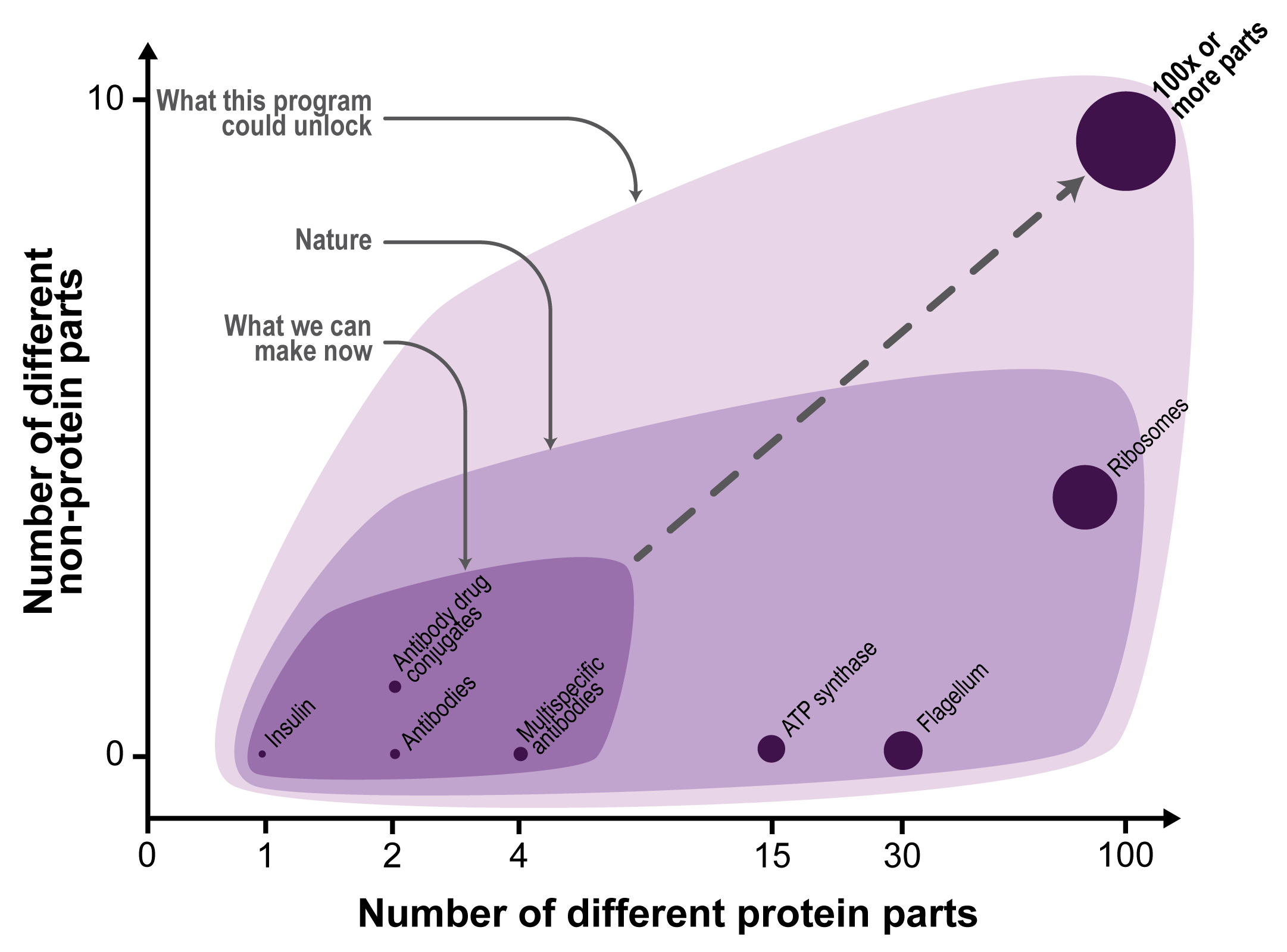
Table of Contents
- Executive Summary
- Who is this roadmap for?
- What is this program trying to do?
- How this program is structured to get this technology into the world
- Elaboration of the research and development phases
- How are proteins designed and synthesized today? What are the limitations of the current approaches?
- Design strategies to make synthetic proteins
- Manufacturing of synthetic proteins
- Modification of synthetic proteins with non-protein parts
- One-pot self-assembly is the status quo to make things from protein
- Synthetic chemistry as done on solid supports
- Summary of limitations of current protein manufacturing processes
- What is technically new in this program’s approach? Why has it not happened yet, why will it be successful now, and why is this approach the way to do it?
- What is technically new in this approach?
- Why has it not happened yet and why is Speculative Technologies needed?
- Other possible approaches for making multi-component protein structures
- Who should care? What difference will this program make?
- Why target the program towards the biopharmaceutical industry?
- Multispecific antibodies where multiple specificities can be encoded
- Protein therapeutics loaded with multiple non-protein parts
- Multivalent and conjugate vaccines
- Prototypes of cell-free chemical factories made with spatially organized enzymes
- Connecting the program to a longer vision of functional nanotechnology
- What are the risks?
- Scalability of the macromolecular additive manufacturing
- Yield of multi step reactions
- Overspecialization of specific building block designs
- Risks imperiling transition of the system to the biopharmaceutical industry
- How is the program structured?
- Phase 1: Solid-phase support material development and protein building block development.
- Project 1A. Solid-phase support material development for large macromolecular building blocks.
- Project 1B. Design and preliminary testing of standardized modular protein building blocks.
- Phase 2: Implementation of protein building blocks and using the solid-phase supports to make multi-component macromolecules.
- Project 2A. Synthesize protein building blocks, anchor them to a solid-phase support, and develop protocols for linking building blocks to one another.
- Project 2B. Demonstrate solid-phase assembly of multi-component macromolecules using established building blocks.
- Project 2C. Demonstrate solid-phase assembly and modification of multi-component proteins with up to ten different parts.
- Phase 3: Continuous flow processes to build and modify large multi-component proteins.
- Project 3A. Development of continuous flow processes for macromolecular additive manufacturing.
- Project 3B. Automation of liquid handling where different structures can be made by programming reagent flow.
- Project 3C. Yield optimization of building block addition and use of the system to make multi-component proteins with up to one hundred different parts.
- What are the benchmarks for each phase of the program?
- Conclusions
- Acknowledgements
II. Who is the roadmap for?
This roadmap describes a process that might make it possible to make nanostructures from proteins with far more complexity than what can be done with the existing approaches. The purpose of this roadmap is to lay out an actionable plan to coordinate people to build this technology and it is intended for the following readers:
- End users who could imagine using the matured technology which could include people working in biopharma, biotechnology, chemical manufacturing, medical and fitness wearables, or beyond.
- Potential builders who could be funded to create this technology, including people from academia, government research labs, start-ups, or established companies.
- Individuals and groups that might be interested in providing philanthropic support of Speculative Technologies to push this forward.
- Or anyone with an interest in the sciences who thinks there should be more ways of putting molecules together to bring useful — or even science-fiction — things to the world.
This roadmap is a living document and needs input from this community. Please reach out to us if you are opinionated about these ideas or can think of ways to make it happen!
III. What is this program trying to do?
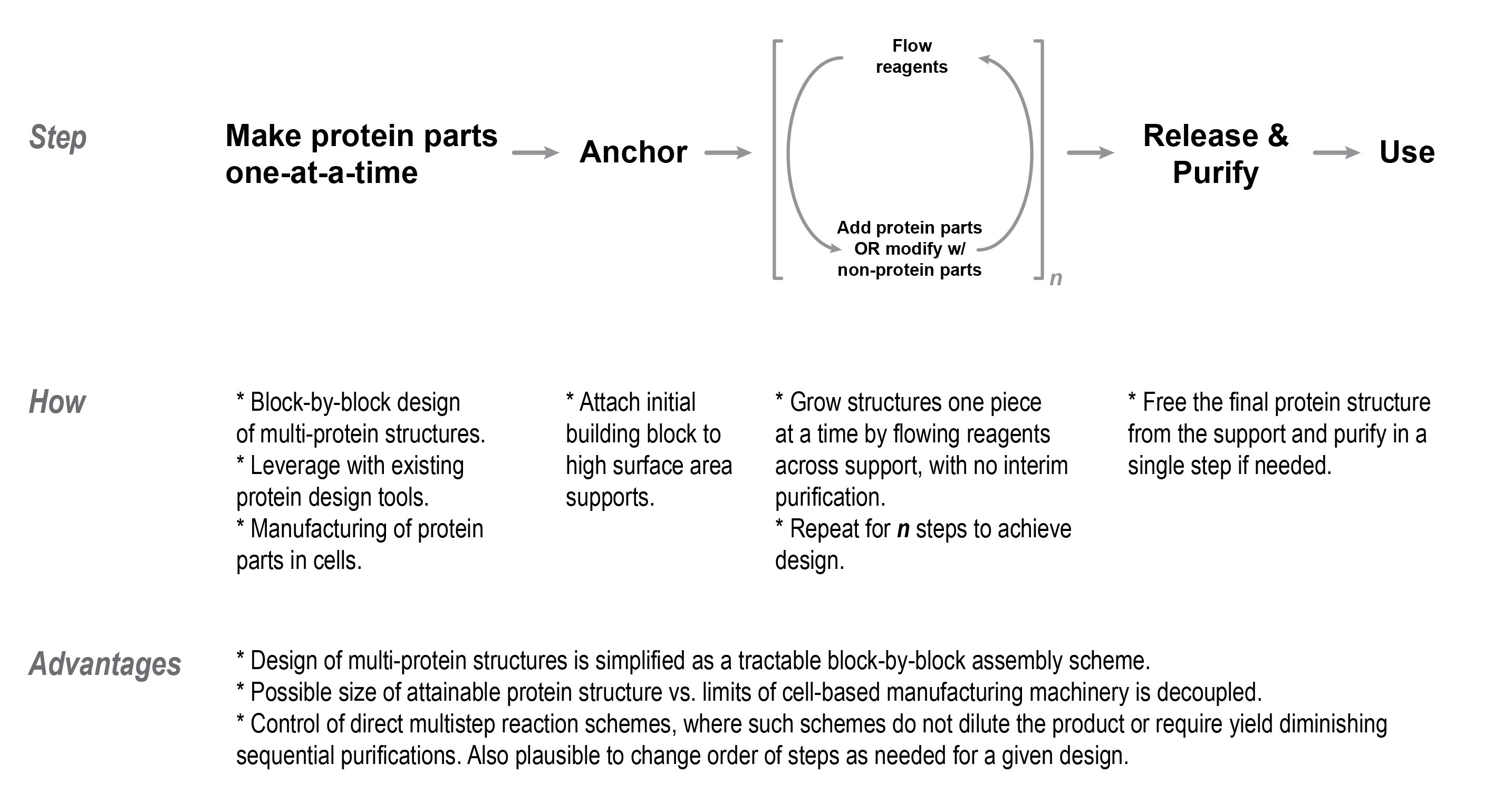
The goal of this program is to devise a new process called macromolecular additive manufacturing to make multi-component protein nanostructures from scratch with tens to hundreds of different parts, as shown in Figure 1.
These multi-component proteins and the infrastructure to make them could unlock new multifunctional therapeutics, vaccines, and enzymatic chemistry. The system could be further adapted to utilize different molecular building blocks, allowing for the assembly of a wide variety of new chemicals and materials with capabilities that are currently hard to imagine.
This program will support research and development of three phases to lay the groundwork for macromolecular additive manufacturing, as shown in Figure 2.
-
Developing solid-phase supports as a ‘workbench’ from which multi-component proteins are grown piece by piece. There is a dearth of support materials for handling large macromolecules at scales that would make them useful beyond the research lab and this is a barrier that needs to be overcome for macromolecular additive manufacturing to flourish.
-
Creating standardized protein building blocks that can be fused together one at a time into multi-component proteins by anchoring them to the solid-phase support.
-
Maturing macromolecular additive manufacturing by integrating the solid-phase supports and protein building blocks into a continuous flow system, where different multi-component proteins are created by programming the order in which liquid reagents are passed through the system.
Macromolecular additive manufacturing is fundamentally different from the existing methods to design and make proteins because scaling from two components to hundreds would be straightforward, depending only on the number of sequential steps.
A. How this program is structured to get this technology into the world
There are two paths this program will use to get this technology into the world:
-
It will incentivize top-leaders in academia and startups to use macromolecular additive manufacturing to build multi-component proteins that have up to one hundred times more parts than what current tools routinely allow. Achievement of this goal would demonstrate the power of the system for making otherwise impossible multi-component proteins. In turn, this would spark a mindset shift to challenge a deeply entrenched assumption in protein self-assembly: That the building blocks themselves must be endowed with the information to orchestrate their assembly into multi-component proteins. The program would give leaders in protein design the tools to make synthetic multi-component proteins of unrivaled complexity. In turn, it would become evident that status quo approaches can be bolstered with solid-phase methods as per this program.
-
This program will tailor the system to build multi-component proteins of intermediate complexity at scales relevant to the biopharmaceutical industry, because such structures could be useful to make the next generation of advanced protein biologics. In turn, there is significant funding in biopharma which will be necessary to carry macromolecular additive manufacturing forward after the completion of this program.
B. Elaboration of the research and development phases
i. Solid-phase supports explicitly for handling large macromolecules
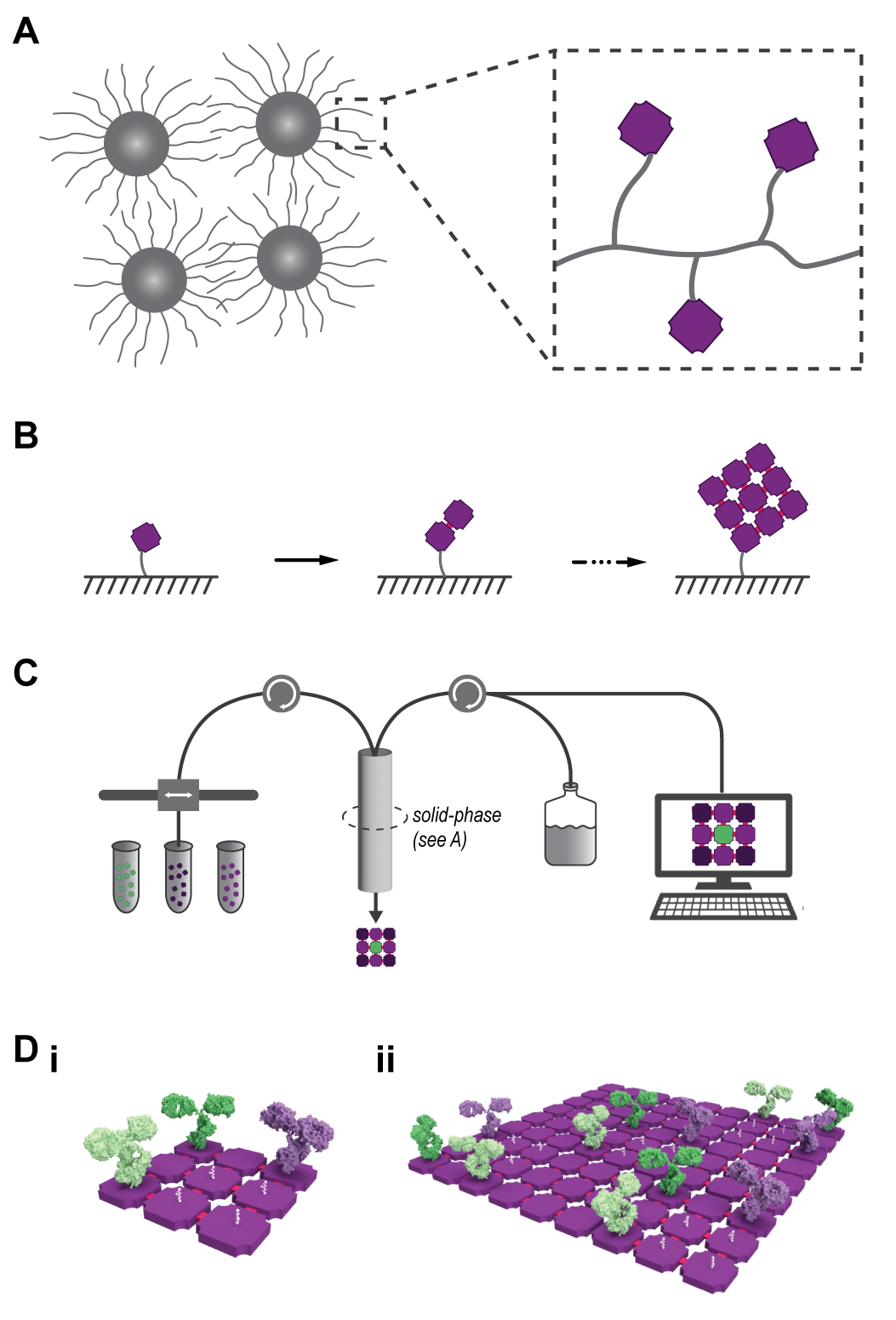
Solid-phase synthesis supports, as inspired from Bruce Merrifield’s Nobel Prize winning invention[2] to synthesize user-defined strings of nucleotides or amino acids, need to be designed for larger macromolecular building blocks. The solid-phase support material — be it in the form of a microscale bead, resin, or polymer network — can be conceived of as a craftsperson’s ‘workbench.’ From the user's perspective, the initial nanoscale building block is anchored to the much larger support material, at which point they may add and remove liquid solutions containing additional building blocks. This process is repeated to grow the structure piece by piece until the full product is achieved. Finally, the user releases the structure from the solid-phase support to use it for whatever downstream application they desire.
The utility of these solid-phase supports is in how they give the user freedom to change the liquid surroundings much more readily than if the product were free floating in solution. This enables the addition of more proteins, non-protein macromolecules, or other chemical groups that cannot be attained using existing solution-phase methods.
However, the solid-phase supports currently available are not suitable for handling large multi-component proteins. As such, this program calls upon chemists, biomolecular engineers, bioprocessing companies, and companies that are immobilizing enzymes for chemistry to develop solid-phase supports with the following properties:
-
Large surface areas for anchoring macromolecules so there is a path to increase the scale of product towards what is attainable using existing protein manufacturing methods. For instance, the surface properties of the material could be modified by grafting polymer brushes increasing their effective surface area.
-
Reusability of the support, so that a batch of the material can be used to create multiple products in continuous process infrastructure.
-
Sufficiently porous, so that liquids enveloping the solid-phase can be readily exchanged. Otherwise, the inability to add the needed reagents could become a yield-limiting failpoint.
-
Ability to match the physical properties of the material to different types of building blocks so as to not cause yield limiting adsorption and aggregation of the building blocks and product.
-
Activatable chemistry so that a protein building block can be stably joined and released from the solid support during the first and last stages of synthesis. This is necessary so that yield diminishing leaching does not occur through the stepwise growth of the product.
ii. Standardized protein building blocks
This program will further develop standard protein building blocks (i.e. small folded single proteins) that can be assembled into multi-component proteins, in a manner akin to plugging toy Lego bricks together. There are two critical benchmarks that must be achieved with these building blocks.
Firstly, these building blocks are intended to demonstrate that macromolecular additive manufacturing can create multi-protein structures composed with up to 100 distinct building blocks. This number of parts would suggest that the system could make assemblies with complexity that begins to rival nature’s protein machines including ribosomes, flagella, and ATP synthases that are composed from dozens of parts. To achieve this, the user could use the solid supports to affix the first building block, and then add additional building blocks one after another until the desired multi-component protein is completed.
Secondly, each protein building block is to feature some sort of binding site so that it can be bound to other cargo. This would enable the synthesis of multi-component proteins, where the attached cargos add new functions to the otherwise structural protein building blocks. The building blocks, assembled into arbitrary multi-component arrays of proteins, could usher in a world of protein nanotechnology that is reminiscent of milestones achieved in DNA nanotechnology. In the near term, such multi-component protein ‘breadboards’ could be used as ‘artificial’ antibodies, protein therapeutics containing several non-protein drugs, multivalent conjugate vaccines, or for prototyping cell-free enzyme factories composed from multiple enzymes.
Some additional properties of the protein building blocks that will be necessary include:
-
Robust, highly specific, and high yielding reactivity for each building block addition. This is challenging, but might be attained by carefully designing interfaces between the building blocks using state of the art protein prediction and design algorithms, as well as matching of the physical properties of the solid support to those of the building block.
-
Triggerable initiation of formation of stable chemical bonds between building blocks as they are added to a growing multi protein structure.
-
Scalable synthesis of the individual protein building blocks. For example, it might be possible to make the constituent parts with traditional cell lines which is appealing because it would leverage existing tools that excel for single proteins.
It is also notable that the solid-phase supports, methods, and infrastructure could eventually enable assembly of building blocks made from non-biological materials, such as peptoids[3], spiroligomers[4], or even inorganic compounds. This would be a path to realizing functional nanomaterials with properties that supersede limitations of biology, such as the development of ultrastable ‘enzymes’ that maintain their ability to make chemicals at high temperatures for long periods of time.
iii. Continuous flow processes integrating solid-phase support and building blocks
This program will integrate the solid-phase supports and building blocks into a continuous flow reactor, where the addition and removal of the reagents can be automated so that it is user friendly to grow multi-component proteins with many constituent parts.
Each growth step might require the user to discard the solution containing the excess unreacted building block from the prior step, replace it with fresh solution with the next building block, and also add any other reagents in the correct order to fuse the building block to the immobilized product. In principle, it would be possible for the user to manually intervene to exchange the reagents on the solid-phase support. However, the user time required to perform such operations would be untenable if the product contained more than a handful of parts.
It will be necessary to get process experts to improve the macromolecular additive manufacturing infrastructure to be able to make multi-component proteins with ~100 distinct parts. The following criteria must be satisfied with the continuous flow system:
-
It must be possible to automate the flow of reagents by programming the order in which they are added on a computer controlled system.
-
The solid-phase support once integrated into the continuous flow system must be robust enough to survive multiple cycles of reagent exchange. For instance, it must not become compacted to the extent that it becomes plugged and unusable.
-
The reactor chamber containing the solid-phase must be able to undergo full reagent exchange such that there are no stagnant zones where new building blocks are unable to contact the support material.
IV. How are proteins designed and synthesized today? What are the limitations of the current approaches?
Let’s start from the most basic level. What are proteins and why are they useful as a macromolecular material?
-
Proteins are nanoscale three-dimensional objects that are folded from linear sequences of amino acids called polypeptides. The sequence of amino acids in the polypeptide determines the shape and properties of the ultimate folded protein, which in turn determines its function. This could include binding pockets that ‘grab’ other small molecules, sites to do catalytic transformations of molecular substrates, affinity domains to bind other proteins, or domains that bind to other copies of the same protein (or a set of different protein subunits) to form larger multi-component proteins that do functions which a single protein could not do in isolation.
-
In the natural context in cells, the instructions to make proteins are encoded in the genome in DNA that gets transcribed to RNA that gets translated into polypeptides. The polypeptides fold to form three-dimensional shapes that become functional proteins. Folding is done by self-assembly[5], which is directed by the intermolecular forces between atoms within the polypeptide, the solvent, and other surrounding molecules.
-
Proteins in biology often do not act in isolation; rather, they are part of molecular systems where they self-assemble into assemblies and associate with other proteins and molecules.
-
Outside of their natural context, people make synthetic proteins to make medicines, vaccines, diagnostics, and enzymes, the latter of which may be used to manufacture chemicals, nutrients, and pharmaceutical ingredients.
-
With medicines, biopharmaceutical companies are making increasingly complex protein therapeutics. This industry has transitioned from merely repurposing natural proteins as therapies towards multi-functional therapies that combine several protein and chemical parts into single entities. In turn, this has enabled new therapeutic mechanisms for fighting cancer, infectious agents, and autoimmune disorders.
A. Design strategies to make synthetic proteins
Design of synthetic proteins, in both research and industry labs alike, has been and remains challenging. The engineer typically relies on discovering, repurposing, and tweaking protein designs from nature to serve the needed application. Realizing such protein designs requires expert artisanal knowledge that is not trivial to acquire.
More recently, there has been advancement of physical and machine learning algorithms, such that protein structure and properties can be predicted from the polypeptide sequence. These modern tools have enabled engineers to create synthetic proteins deviating more drastically from natural counterparts over shorter timescales, however robust capabilities to rationally design de novo proteins are still a work in progress. Machine learning models are only as powerful as the structural data upon which they have been trained, which limits the extent to which new proteins without a natural counterpart can be designed.
These protein design algorithms are even more limited in their ability to make larger multi-component proteins with more than a couple distinct proteins. The latest prediction tools are adept at making protein lattices and virus-like protein shells that are composed from symmetric arrangements of one or two different protein subunits. By contrast, natural protein machines including ribosomes, flagella, and ATP synthases are composed of dozens of distinct protein subunits that are arranged asymmetrically (see Figure 1). The inability to make large multi-component proteins is because design algorithms are not able to create the large numbers of compatible (i.e. orthogonal) protein-protein binding interfaces necessary to adjoin the many different protein parts.
Irrespective of how the initial design is done, users must iteratively test the designs to see if cells can produce them and whether the product performs the desired function. It is hard to know whether a new protein design will form successfully into the desired structure matching the design. Cells might struggle to produce the well formed protein resulting in the formation of insoluble aggregated junk. Nuances including the codons selected to encode a protein, number of protein subunits, and biophysical properties of the protein can have non-obvious and subtle effects on the yield. In turn, this narrows the accessible protein design space to only what cells are capable of manufacturing.
B. Manufacturing of synthetic proteins
Synthetic proteins are typically manufactured using living cells that are reprogrammed to make them. DNA instructions encoding the protein are added to the cells, some of the cells are added to a nourishing broth that lets them proliferate, and the cells are collected after sufficient time has elapsed for ribosomes to produce the protein. The volume of cell ‘culture’ required is determined by the amount of product needed. Testing and optimizing the protein in the laboratory might require ~10-3 liter batches of cells, while production of a therapeutic protein in the biopharmaceutical industry is often done in batches of ~104 liters to make kilograms of the product.
There are many different species and cell strains to choose from to make proteins, with each having unique properties that must be matched to the requirements of the particular product. One parameter for cell selection is whether there are ‘post translational’ modifications that must be added to the protein for its desired application. After ribosomal synthesis, certain cell lines have specific machinery to add chemical modifications to the protein, including the addition of sugars, internal chemical bonds, or other non-protein molecules. These modifications have proven particularly important for therapeutic antibodies, where certain cell lines (e.g. bacteria) are not equipped with the machinery to add sugars necessary for the therapeutic to not be destroyed by the immune system.
One limit of protein manufacturing in cells is that the length of polypeptide — and therefore the size of the ultimate protein — is limited[6]. As such, formation of larger proteins requires designing smaller protein parts that self-assemble into bigger structures. It should also be noted that different cell strains and species for making proteins have unique constraints which may make them challenging to perform consistently in large scale manufacturing. For instance, the mammalian Chinese hamster ovary (CHO) cell line for making large amounts of therapeutic antibodies is fussy towards small variations in temperature and concentration of carbon dioxide. This is in stark contrast to other prokaryotic cell lines (e.g. Escherichia coli) that are more robust to making protein products, even in the presence of such environmental changes.
The user must collect the protein once it is manufactured by the cells. This could be achieved by either triggering the cells to secrete the protein using special cellular machinery, or by gathering and breaking the cells open to free the protein. Purification may be required to separate the product protein from other contaminants, with the stringency of purity being especially important for biopharmaceutical applications. One persistent challenge is that purification tools — whether it be using chromatography, membrane selection, or affinity matrices — are cumbersome, expensive, and will lead to inevitable loss of product that will limit yield.
C. Modification of synthetic proteins with non-protein parts
After purification from cells, the user may modify and append the proteins with non-protein parts. This has been particularly important for therapeutic proteins, where modifications such as the attachment of a chemical to kill cancerous cells, might be a critical part of the functional drug. To add these, the protein is mixed with the cargo and other reagents to activate bonding sites and link them together. The number and order of reagents that must be added depends on the chemistry linking the protein to its cargo, with these processes resembling workflows from synthetic chemistry. Additional purification steps may be needed to isolate the modified proteins from the reagents and unreacted proteins, with the degree of purification being dependent on the application of the product structure.
Additional reaction steps and purifications are challenging because it will diminish the final yield of the product. Moreover, there is only a limited set of orthogonal and compatible linkage chemistries to add other cargos on a protein structure[7]. It becomes untenable to sequentially add several modifications, further limiting the design space of proteins that can be manufactured at scale. For instance, while there are many examples of protein therapeutics that attach a single drug or polymer, there are no therapies on the market that have more than one type of modification on a single protein.
D. One-pot self-assembly is the status quo to make things from protein
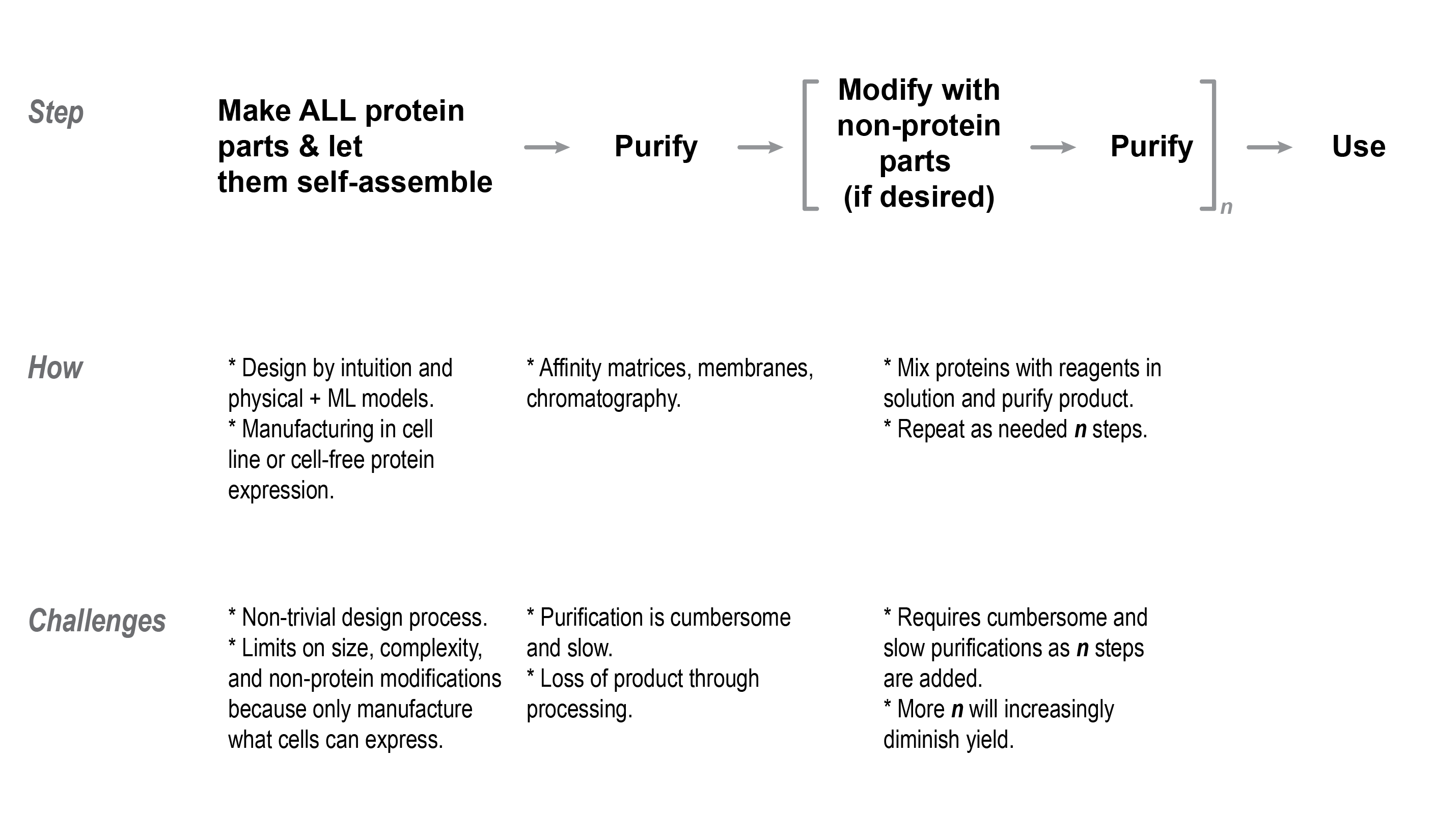
The aforementioned approach of making proteins in cells, as well as other processes to make macromolecular structures from other materials, create the final product by mixing the building blocks together to let them “self-assemble.” This is a tacit assumption about processes in biology and the macromolecular sciences and needs to be highlighted to explain the value of this program. The reaction vessel could be a tube where the composition of the reactants is dictated by the user or the liquid environment inside a cell. The building blocks move around in free solution and bump into one another and specific regions on the constituent components stick and engage to self-assemble into a multi-component structure like pieces of a puzzle.
The way in which these binding domains are integrated into the constituent pieces is a big challenge in protein design, where the engineer is encoding an algorithm into the building blocks that ‘tells’ them how to fit together. To add to this challenge, they must also incorporate other functions — such as the ability to bind to the site of disease for a therapeutic protein — in the design. Optimizing the assembly algorithm and other functionalities together is challenging because it demands structural complexity of the constituent molecules, may be hard to rationally engineer the protein to properly self-assemble, and creates failure points in which monomers can mis-assemble.[8]
E. Synthetic chemistry as done on solid supports
Traditional synthetic chemistry involves mixing small molecular-scale reagents, as similar one-pot self-assembly. However, they are different because the smaller molecular building blocks in synthetic chemistry carry less information to direct their formation into products. Hence, the molecular structures possible by mixing all the reagents in one-pot are less complex compared to those from self-assembly of proteins. Increasing the complexity of products with chemistry of molecular building blocks requires building the structure up by sequentially doing simpler steps one-after-another.
One particularly enabling synthetic approach to make complex products from simple molecular building blocks is solid-phase synthesis. The approach was pioneered by Bruce Merrifield and won the 1984 Nobel Prize in chemistry for how it allowed chemical synthesis of long polypeptides[9]. In the method, an amino acid building block is linked to a solid resin, immobilizing the building block with respect to the surrounding liquid solution. Next, the user adds a reagent to activate one end of the immobilized product to make it reactive to polymerize it when the next amino acid is added. This process is repeated until the desired peptide is constructed, at which point it is cleaved from the resin to collect the product. This methodology is ubiquitous for the synthesis of nucleic acids, peptides, and other chemicals that are used in research labs. Moreover, the technique is also used to make drugs that are composed of short peptides.
Notably, there have only been limited attempts to apply solid phase processes for assembly and modification of larger macromolecules, with limited demonstrations to modify proteins in laboratories[10]. However, the methodology has not been applied for large scale production of macromolecular products.
F. Summary of limitations of current protein manufacturing processes
The current methods for making proteins are fraught with limitations at all levels of design and manufacturing that bounds the complexity of synthetic proteins. Design algorithms struggle to make multi-component proteins composed from many unique parts and it is unclear how long it would take for these in silico tools to improve to structures which rival the complexity of natural proteins. Existing manufacturing relies on ribosomal protein synthesis, but cellular machinery is limited in the size of proteins that can be synthesized, is subject to non-obvious nuances of protein expression and folding, and is restricted in the range of post-translational features that can be added with cellular machinery. Moreover, downstream purification and chemical modification processes of proteins can be clunky, where yield limits of such processes make it difficult to add more than a single modification to a given protein. Finally, there is a gap between state of the art solid-phase chemistry and the self-assembly methods for making proteins in solution. This program intends to merge processes from each of these worlds so that even more intricate protein structures can be attained.
V. What is technically new in this program’s approach? Why has it not happened yet, why will it be successful now, and why is this approach the way to do it?
A. What is technically new in this approach?
The key technical differences of this program versus existing methods to make proteins, and practical consequences of these differences, are as follows:
- Most of the steps to build up a protein structure with multiple parts will be done on solid-phase supports, in contrast to existing methods which are done in solution. Anchoring the product to a solid-phase support enables unprecedented control over the steps to make intricate proteins. The fluids containing the input materials to build up the protein can be easily exchanged one after another, while simultaneously mitigating the loss of product that would otherwise happen if the steps were carried out entirely in solution.
-
Use of solid-phase supports to build up proteins would establish a fundamentally different process to manufacture macromolecular structures versus the status quo. It would become possible to do cycles of reagent exchange, allowing the sequential addition of protein and non-protein parts without having to do unwieldy processing steps to recover intermediate products. Once the full structure is assembled with all the protein and non-protein parts, it is released from the support with only one purification step to recover the product. By contrast, reagents cannot be easily exchanged using processes done entirely in free solution. This is problematic for doing sequential addition of parts to a protein because reagents between reaction steps might be incompatible with one another. Moreover, the product structure might become diluted to the extent that it becomes difficult to recover without noticeable loss in downstream purification steps.
-
The macromolecular additive manufacturing system can be adapted to work with materials produced using traditional solution-based methods, enabling the engineer to choose the best approach for making a given segment of the multi-component protein. For instance, the protein parts (e.g. standardized protein building blocks or other protein cargo) could be synthesized in cells to leverage how such tools can make large amounts of a single protein. The protein parts could be gathered from cells and assembled sequentially into larger multi-protein structures using solid-phase supports, allowing the user to access proteins that cannot be made in cells. Moreover, the control offered with solid supports could be used to add multiple different non-protein modifications using a single linker chemistry by strategically splitting protein structure into multiple pieces. Each piece could feature a reactive residue that binds different cargos using the same chemistry. Different cargos could be added to each piece individually in separate reactions, at which point the two protein pieces could be assembled into a single unit using solid-phase supports. In effect, pseudo orthogonality could be obtained from a small set of linkage chemistries.
-
The solid-phase support system would be amenable to both batch and continuous flow processes to add and remove reagents. The user could manually add reagents to a batch of the solid-phase support, and this would be useful for creating assemblies with a small number of parts or for preliminary testing and optimization. Such batch procedures are typical of protein production and modification processes for laboratory and industrial manufacturing of proteins. Alternatively, the user could add reagents using a combination of fluid handling pumps and continuous flow through the solid support. This approach is desirable because it would be possible to program and automate reagent flow, so that the user could create larger assemblies composed of dozens of parts. It would be possible to miniaturize reagent use to make small amounts of product for prototype testing using microfluidics. At larger scales, it could be possible to conserve reagent use by recycling unreacted materials between assembly steps.
- The algorithm to make higher order multi-component proteins would be encoded in the order that building blocks and reagents are exchanged across the solid-phase support to “grow” the anchored product. By contrast, current approaches in protein design use one-pot self assembly, where the algorithm to make multi-component proteins is contained in the building blocks themselves.
- The system could make it simpler to design and assemble multi-protein structures. It would become possible to use standardized building blocks — such as proposed in this program — where the process of fitting one block to another is highly optimized. Downstream users of the system could program the flow of these building blocks to make new multi-component proteins. For instance, one could build asymmetric arrangements of dozens of distinct building blocks, which are untenable with the existing design tools and one-pot self assembly protocols.
B. Why has it not happened yet and why is Speculative Technologies needed?
Although many of the fundamental pieces for macromolecular additive manufacturing have been in existence for decades, including solid-phase supports and recombinant protein engineering, the technology has not matured beyond limited demonstrations at small scales in isolated laboratories. There are several reasons why this might be:
-
The ideas in this program challenge deeply ingrained assumptions tacitly held by experts in biology and macromolecular engineering who think about processes to make things from proteins. These people typically make synthetic proteins by expressing them in cells, where ribosomal synthesis and self-assembly elegantly puts the proteins together into the product. They generally do not have to think about the nitty-gritty stepwise details of how the structure comes together. The idea of putting proteins together in several distinct stages, with each requiring some degree of intervention, is a mindset that is reminiscent of synthetic chemistry. This program and where Speculative Technologies can excel is to incentivize pragmatic merging of both approaches.
-
The approach for this program requires combining several modules into a functioning system, which requires input from siloed research disciplines such as polymer chemistry, process chemistry, and protein engineering. Cross-disciplinary collaboration and integration of systems is difficult to sustain and fund among academic investigators. Consequently, examples of where researchers have applied solid-phase methods to construct and modify protein structures have been limited to niche examples[11] that have been of little impact beyond the individual investigator. For instance, better chemistry is needed to develop solid-phase supports explicitly for large macromolecules. Researchers are currently stuck using materials from commercial vendors. These materials may cause aggregation of large proteins because they are optimized for use with smaller peptides, or else have limited surface area that dramatically limits the scale of product which may be attained. However, a coordinated research program with Speculative Technologies could incentivize collaboration between polymer chemists, companies that make resin materials, and companies doing immobilization of enzymes to make scalable solid-phase supports for macromolecular assembly. Moreover, process chemistry is needed to make this system usable to build proteins with many components. Examples of where solid-phase supports have been used to modify proteins or build up other macromolecules[12] are done as batch processes. This is suitable for preliminary development of these approaches, but requires too many time intensive user interventions to be practical for making larger products. Continuous flow processes would be necessary to make proteins of the complexity that would demonstrate the value of this system. This could be attained by incentivizing collaboration between macromolecular engineers, process chemists, or other groups that have access to costly platforms (e.g. solid-phase peptide synthesizers) that could be adapted to test processes at the pilot scale.
-
Protein design tools have not been sufficiently generalizable or user friendly to make multi-component protein structures. Forward design of proteins has required niche knowledge that is held by a handful of research groups and industry experts. Only recently has there been marked advancement in the ability to do forward design of protein structures with folding prediction tools based in machine learning and large language models. We are at a cusp where these increasingly capable tools could be leveraged to make robust input materials — such as protein building blocks that can be activated on command to covalently bond to one another — for this system.
-
Incumbent protein manufacturing infrastructure in the biopharma industry is embedded and hard to challenge. Development of protein therapies is a risky endeavor, with only about ~12% of the drugs entering the development pipeline successfully making their way to full approval needed for the drug to enter the market. The full research and development cost of a given drug that successfully passed full approval was in between $1–2 billion US dollars[13]. Successfully developing a new protein biologic using methods akin to macromolecular additive manufacturing will require testing and scale-up of new processes that would add further risk to an already expensive and risky endeavor. In turn, this would dissuade startups and large companies from taking on this work. For instance, resource strapped biopharma startups would find the fundamental development of macromolecular additive manufacturing to be a distraction from the core goal dictated by investors: that they must discover drugs that are safe and add value with a noticeable therapeutic effect for treating a disease or disorder. Startups have to make the transition from discovery of drug candidates to production of larger quantities of the therapy for different stages of the approval process in clinical trials. In turn, this might necessitate them to seek external help from contract development and manufacturing organizations (CDMOs). However, it would be difficult or costly to find such external partners with the capabilities to make protein therapies requiring non-existent manufacturing tools that do not have a FDA current good manufacturing (cGMP) precedent.
C. Other possible approaches for making multi-component protein structures
There are other potential approaches for building multi-component proteins versus macromolecular additive manufacturing. Other plausible approaches and their disadvantages — which point toward the approach in this roadmap — are listed below.
-
We could wait for protein prediction and design tools to improve to the point where they can make any arbitrary protein. Algorithms for predicting protein folding have been rapidly improving[14] because of deep learning and large language models. These developments will inevitably also make it easier to make multi-component proteins too. However, the timelines are unclear as to how long it will take for these tools to be reliable and user friendly to realize this. There could be unforeseen challenges to design adjoining interfaces of proteins for them to successfully self-assemble into the product. For instance, the structural DNA nanotechnology community — despite DNA being a much more tractable and easy to design versus protein — has shown that misbinding of parts limits complexity despite care in tuning the interactions between the parts[15]. Moreover, there will still be fundamental process limitations in cell based manufacturing of proteins, regardless of how capable in silico prediction and design software becomes.
-
It might be possible to develop a large set of orthogonal protein binding domains, where strategic placement of different domains on the various constituent parts would let them self-assemble into multi-component proteins. Nonetheless, it will be challenging to perfect binding of all the various domains so as not to make malformed side products. It might become necessary to split reactions up into multiple steps[16] to lessen such yield depleting erroneous interactions. In turn, this will make it unwieldy to scale these approaches to proteins with more than a handful of components.
-
Protein building blocks could be placed into multi-component protein structures using templates to hold building blocks in such a way that allows them to be fused together. The templates could be made from easy to engineer materials like DNA origami to circumvent challenges in designing less tractable materials like protein. One possibility could be to attach the DNA nanostructures to the solid-phase material to functionalize the supports and enable them to direct arrangement of the protein building blocks. However, this approach would require a unique template to be tailored for each new design, which could be unwieldy if trying to prototype many multi-component proteins. There could be challenges in making the templates compatible with the building blocks, where there might be geometric mismatch of the template to the extent that it is unable to fuse the proteins together.
-
It might be possible to build macromolecular machines to ‘print’[17] or do ‘positional’ chemistry of building blocks. This approach is fundamentally different from the approach here; it would be akin to building something like an artificial ribosome to stick macromolecular building blocks together and would be a huge step towards functional nanotechnology. However, it would be far and away the most complicated approach since it necessitates constructing several advanced nanoscale components that must simultaneously work together as a single macromolecular machine.
Finally, it should be noted that work that could be done towards the alternative approaches need not be exclusive of the macromolecular additive manufacturing program here. The solid-phase support system as per this roadmap could be combined with any of the approaches above to make it easier to do reagent handling to make even more complicated multi-component proteins.
VI. Who should care? What difference will this program make?
The goal of this program is to make it easy to design and build multi-component proteins. This would unlock new infrastructure to build a breadth of intricate synthetic proteins that could enable:
- A. The biopharmaceutical industry for making protein therapies and vaccines.
- B. Prototypes of cell-free chemical factories made with spatially organized enzymes.
- C. Connecting the program to a longer vision of functional nanotechnology.
A. Why target the program towards the biopharmaceutical industry?
There are two reasons biopharma is an important downstream anchor for macromolecular additive manufacturing.
Firstly, this program would continue an emerging trend in modern biologics: Protein therapies and vaccines are mixing and matching increasing numbers of protein and non-protein components. State of the art therapies such as multispecific antibodies combine up to four (or a slightly more) distinct protein parts[18]. In turn, this allows multi step treatment mechanisms to be encoded within the therapy.
However, the upper level of product complexity is impeded by what can be made within cells, unwieldy design tools, and clunky downstream processes for adding non-protein modifications that have no biological precedent. The biopharmaceutical community necessarily focuses on proteins that may be made with cellular manufacturing tools, with the unsaid premise that pursuit of designs beyond what these tools can make is futile. Consequently, macromolecular additive manufacturing could make multi-component proteins that could significantly expand the design space for therapies and vaccines.
Secondly, there are significant resources in biopharma which could help carry macromolecular additive manufacturing forward after the completion of this program. For instance, venture capital dollars could embolden builders of the technology to launch a startup to create a multifunctional protein drug using the system. Alternatively, it might be possible to get developers of the specialized solid-phase supports to sell these materials directly to biopharma companies so that the industry could adapt them to internal processes.
The three subsections below explain more about the state of the art of current therapies, limitations of what can be made, and how macromolecular additive manufacturing could circumvent these barriers.
i. Multispecific antibodies where multiple specificities can be encoded
New antibody therapies called multispecific antibodies bind two or three different biological targets. This allows developers of biopharmaceuticals to create therapies that can simultaneously block multiple signaling pathways and immune checkpoints. They may also make medicines which engage targets on the site of disease with the other targets on cells in the immune system, making it possible to induce formation of protein complexes that eliminate diseases otherwise invisible to the body. Hence, multispecific antibodies allow encoding of nuanced multi step treatment mechanisms, which is a notable advance over natural antibodies binding to only a single target.
However, the existing multispecific antibodies are limited to tweaks of naturally occurring proteins and must ultimately be manufactured using cells. This includes the addition of shape complementary ‘knob-in-hole’ features on antibody subunits, or concatenating genes to make chimeric fusions of antibody fragments. Such approaches drive the constituent pieces to self-assemble by using the existing cellular manufacturing tools to make them. However, the process of designing and optimizing these modified proteins is highly specialized, generally restricted to single therapies on a case by case basis, and limited to the constraints of what cells can manufacture. This means that the size of the proteins, the number of different targets they can engage, and the range of encodable therapeutic mechanisms are restricted.
The macromolecular additive manufacturing system would allow modular plug and play assembly of dozens of protein building blocks that can position therapeutically useful cargo, without the constraints of cell-based protein manufacturing. It may be possible to create multi-component proteins that function as synthetic multispecific ‘antibodies,’ capable of engaging with dozens of targets simultaneously. This would open a vast design space for developing the next generation of therapeutics.
ii. Protein therapeutics loaded with multiple non-protein parts
Protein biologics may be functionalized with other non-protein chemistries after initial production of the protein in cells. For instance, enzyme therapies may be appended with polymers (e.g. polyethylene glycol) to reduce immunogenicity and boost their circulatory lifespan, or an antibody may be conjugated to a drug, wherein a chemotherapeutic that is too harmful to be administered on its own can be delivered precisely to the site of disease. Henceforth, such non-protein modifications add new functionalities to proteins to create new treatment mechanisms.
The current manufacturing processes — with the orthogonality of chemistries for adding parts and difficulty of product recovery — do not lend themselves to adding more than a single non-protein part. The protein is initially synthesized and purified from cells, with non-protein modifications requiring reagent exchange to activate protein linkage sites. Recovery of the protein requires it to be purified from unreacted reagents reducing its final yield, with the yields diminishing further if more purification steps are added. Additionally, finding multiple compatible chemistries to add different modifications is challenging.
Macromolecular additive manufacturing addresses these challenges by immobilizing proteins on a support, similar to securing them in a vise on a workbench. This enables the user to exchange reagents one after another to activate chemistries protein for attachment of non-protein parts, without loss of the anchored protein. Similar chemistries could be used to add different non-protein parts to separate protein building blocks and subsequently linked into a larger entity on a solid-phase support. As such, a minimal set of chemistries could be used to attach many non-protein parts to a multi-component protein. Moreover, the orientation of protein binding to the support could be used to occlude otherwise active residues facing the support, allowing for selective activation of the remaining exposed reactive residues to further enhance control over where modifications occur.
iii. Multivalent and conjugate vaccines
A persistent challenge in vaccine development is that there are often many unique strains of viral and bacterial pathogens, each requiring different versions of the vaccine. This requires development of multivalent vaccines, where the formulation is a mixture of multiple molecules that each display features (i.e. antigens) unique to a particular strain. Another challenge is that the various antigens on these pathogens to which a vaccine may be developed are sometimes only weakly recognized by the immune system and offer little protection. One work around has been to create conjugate vaccines, where weakly recognized antigens are tethered to antigens which engage the immune system more strongly. However, the extent to which the active molecules in vaccines can be engineered to tether multiple components to trigger the immune system more fully against a broader set of pathogens is limited.
The macromolecular additive manufacturing system would make it possible to create scaffolds from protein building blocks, where each building block could be fused to different components. It would become possible to build advanced multivalent vaccines, where the active vaccine ‘molecule’ triggers immune response to multiple pathogen species, potentially lessening the number of vaccinations required to gain immunity to common diseases. The system would make it possible to reconfigure existing vaccine designs to tether additional components, making it possible to adapt an existing vaccine to target emerging pathogen strains. Moreover, the ability to conjugate dozens of components could make it possible to trigger robust immune responses across multiple antigens on a single pathogen.
B. Prototypes of cell-free chemical factories made with spatially organized enzymes
The multi-component proteins in this program could also be used to create prototypes of cell-free factories to synthesize chemicals using spatially organized enzymes, where tens of different enzymes are integrated on otherwise non-functional protein scaffolds. Biological systems — such as polyketide synthases where the order in which synthetic modules are strung together leads to different products — proves that organization of macromolecular parts gives rise to otherwise unattainable behaviors if the parts were just stochastically mixed together. However, there are only limited examples of synthetic systems that robustly exhibit such behaviors. For instance, co-localization of enzymes have demonstrated interesting effects using low concentrations of reagents that are typical of laboratory demos, but when the concentrations of reagents is increased to what might be typical in an industrial reactor the effect of co-localization is much less clear.
There are two particular challenges that have kept these innovations from proliferating beyond the lab. Firstly, it is difficult to build synthetic systems of spatially organized enzymes because of the limitations of status quo protein design and manufacturing. Consequently, academic researchers rely on established protein scaffolds that are often borrowed from biological designs to create these systems. It takes a long time for them to make a new design, it is not possible to string together many pieces, and there is only limited spatial control over how they attach to one another.
Secondly, it is not crystal clear what designs of such spatially organized systems might be more robust and useful for industry. There are many non-obvious microdomain effects[19] that could be rationally engineered to improve the flux and efficiency of such systems. For instance, the charge properties of other macromolecules to coordinate enzymes could be tailored to ‘trap’ product intermediates using reversible interactions such as hydrogen bonding and Van der Waal forces. There are also models that suggest there could be more clever ways such as compartmentalization[20] to build such systems. However, these will remain “future things that should be tested” until there is a way to physically realize them.
Another endpoint goal for the program could be to use macromolecular additive manufacturing to rapidly prototype systems of spatially organized enzymes. In doing so, it could be used to determine potential designs that are useful for making molecules for the chemical and pharmaceutical industries. We could further envision how such a system could be applied to structured competition, where some set of “holy grail” needs for the sustainable enzymatic chemistry industry could be used as a target for participants operating the platform.
However, this goal might risk overspecializing the macromolecular additive manufacturing system to a single use case with spatially organized enzymes. Furthermore, the scales of product which would be required for adoption by chemical manufacturers would be much larger compared to other contexts of use in the biopharmaceutical industry. Nonetheless, this could be a future direction of macromolecular additive manufacturing and another step to follow the aforementioned program goal.
C. Connecting the program to a longer vision of functional nanotechnology
Another goal around which this program could be centered is to use it to enable functional nanotechnology. For instance, the early conceptions of molecular assemblers and Drexler nanotechnology[21] have arguably stagnated and not yet come to fruition. There are three demonstrations which could move work of this theme forward:
-
The system could be designed to make it easier to operate artificial nanomachines that already exist. For instance, there are machines that are built from DNA where motion of parts within each machine are driven by DNA strand displacement[22]. One challenge with these systems is that continued motion requires continued exchange of fluids containing DNA oligonucleotides. Alternatively, such machines could be mounted to the solid-phase supports with a continuous flow reactor serving to exchange reagents as needed.
-
In its essence, the macromolecular additive manufacturing system makes it possible to arrange simple building blocks in multi-component structures that could not be attained by the constituent parts acting alone as per self assembly. This ability is an unstated underpinning of ‘molecular assemblers’ and could be demonstrated with protein building blocks. Moreover, it might be possible to add non-biological building blocks to use the system to build molecular machines more reminiscent of Drexler nanotechnology.
-
The program could be a testbed for demonstrating ‘positional chemistry’ which remains an elusive and important milestone in functional nanotechnology. For instance, the designs for protein building blocks could be refined so that covalent linkages are only induced once a complementary protein ‘tool’ that binds to the interface of adjacent building blocks is introduced.
Structuring this program around this goal could build and engage a community dedicated to moving functional nanotechnology as a mainstream research discipline. In turn, these demonstrations could derisk future programs centered on molecular assemblers with either Speculative Technologies or other government and defense funding organizations. However, this goal is risky because there is not a clear context of use for any of the demonstrations, which could lead to niche results that are ignored by the broader world.
VII. What are the risks?
A. Scalability of the macromolecular additive manufacturing
Solid-phase support materials for multi-component proteins might be difficult to scale for making large amounts of product, which will be necessary for the method to be adopted beyond lab demonstrations.
One particular challenge is the amount of product that may be synthesized from a given volume of reactor is constrained by the surface area of the support material. However, there is evidence suggesting that larger scales could be attainable:
-
Biopharma and biotechnology already makes large amounts of polypeptides[23] and oligonucleotides using solid-phase chemistry. It shows that non-trivial products can be made at scales relevant for industrial applications, even if these macromolecular products are smaller than the multi-component proteins in this program.
-
There are already laboratory-scale examples of where solid-phase chemistry has been used to make macromolecules as large as single proteins, where the scale of protein made per reactor is impressive — this work[24] used a commercially purchased solid-phase support[25] and reported loading it with ~0.5 millimoles of protein per gram of support. If we account for swelling of the support, one mole of protein could be produced in a ~1000-fold smaller volume compared to the volume that would be needed if it were made using the existing methods. Of course, this volume reduction would require high yield of product, which is difficult but potentially solvable.
-
The challenge of limited surface area of the support material could be overcome with fundamental development of solid-phase supports. There are companies that are building increasingly advanced solid-phase supports. For example, leading enzyme immobilization companies[26] are using microparticle supports grafted with customizable polymer brushes that grip the protein while maintaining their function. There are also examples in academia of reconfigurable polymers[27], which may be triggered to condense into a solid to recover nucleic acids where there are no scale-limiting surfaces per se. This program is trying to encourage development of solid-supports that are purpose built for large multi-component proteins.
There is also quantitative evidence to suggest that the scalability of macromolecular additive manufacturing might be competitive with status quo cell based methods for making proteins. It should also be noted that potential downstream users of multi-component proteins from this system require different amounts of material. For instance:
-
The scales required for biopharmaceuticals are dependent on the design and particular use of the therapeutic. A single dose of a traditional antibody is ~1 g, versus a dose of an antibody conjugated to a cytotoxic drug that is perhaps ten times less because of its increased potency. Vaccines typically require an even smaller amount per dose (e.g. ~0.001 g) because of their role in triggering the immune system. There is a thousand-fold range of therapeutic amounts needed among these examples alone!
-
The scales that might be required to prototype chemical factories from spatially organized enzymes could be even lesser yet because it would be reserved for laboratory scale experiments which could be performed in small volumes.
Taken together, there is a path to increasing the amount of product that could be manufactured on a solid-phase support and variety of applications that would require different amounts of product. It is conceivable that as macromolecular additive manufacturing matures that product scale could be increased for applications in biopharma or prototyping of spatially organized enzymes.
i. Quantitative analysis of scalability of macromolecular manufacturing
The scalability of macromolecular additive manufacturing is considered more quantitatively by comparing the estimated space time yield versus the status quo of making antibodies in mammalian cells, as shown in Figure 5. ‘Space time yield’ refers to the amount of protein which may be manufactured in a given volume per period of time. In effect, it is a measure of the productivity of each manufacturing approach. Details of the calculations are explained further in a footnote[28].
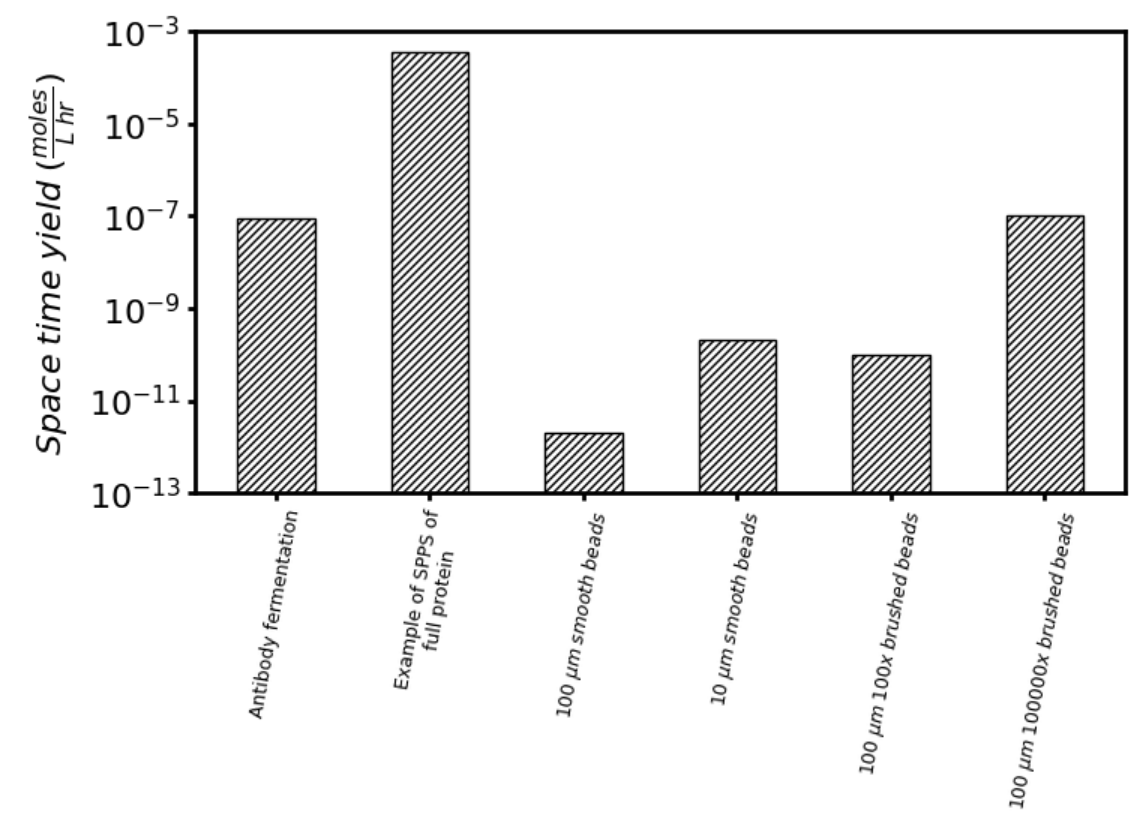
As a reference, an exemplary Chinese hamster ovary cell line might produce 5 grams of antibody per liter of cell culture over 16 days. This corresponds to a space time yield of ~100 nmole L-1 hr-1. As further reference, this previous work[29] used solid-phase peptide synthesis to make a protein (i.e. fibroblast growth factor 1) and was able to achieve a space time yield of ~100 000 nmole L-1 hr-1. This is about a ~1000 fold increase in space time yield versus status quo cell-based methods for making proteins — these are real results, they suggest that remarkable productivity can be attained using solid-phase methodologies, and are inspiring for this program.
The aforementioned observations are interesting, but they only consider proteins as complex as an antibody. What if we approximate space time yield for macromolecular additive manufacturing of a multi-component protein with nine pieces, like the sample structure shown in Figure 2?
Computing this estimate requires a simple model of possible solid-phase supports. We will assume that each building block added to the solid-phase support is attached to the product relatively quickly (i.e. at a rate of ~105 M-1 s-1) and that the liquid solutions of building blocks input to the system are at a moderately high concentration (i.e. ~100 nM). Let us consider the following solid-phase supports:
-
Large 100 µm diameter smooth beads, where the surface is densely packed with the multi-component protein. The diameter of these beads is about the thickness of a sheet of paper and space time yield is ~100,000 fold lower compared to status quo methods to make a mole of product. This is dismal and not promising.
-
We could decrease the size of the bead to increase the surface area for loading product using small 10 µm diameter smooth beads, where the surface is densely packed with the multi-component protein. Space time yield is ~1000 fold lower compared to the status quo. This is more promising, but these beads would be very tiny and this could cause other problems, such as becoming over-packed and unable to let reagents easily pass.
-
There are other strategies where we could engineer better solid-phase supports with higher loading capacity. For instance, the large 100 µm smooth beads could be grafted with polymer brushes that increase the loading of multi-component protein by 100x versus the smooth bead. Space time yield here would similarly be ~1000 fold lower compared to the status quo.
-
Finally, we could further improve this polymer grafting strategy. For instance, the large 100 µm smooth beads could be grafted with polymer brushes that increase the loading of multi-component protein by 100,000x versus the smooth bead. Space time yield here would be comparable to the status quo!
These quantitative considerations suggest that solid-phase supports could be engineered to make large numbers of multi-component proteins in a small space relatively quickly. In turn, it might be possible to develop macromolecular additive manufacturing at competitive scales for certain applications.
B. Yield of multi step reactions
The yield of multi step chemical reactions, where n is the total number of reactions steps and x is the yield for each step, is determined as:
yield = xn-1
Hence, the x yield for each step where a building block is added must be very high in order to have a reasonable yield of the total product. Let’s assume we wish to grow a multi-component protein with 100 parts (i.e. 100 steps). If there is 95% success rate that a given building block is added, then there will be less than a 1% yield of the product. However, if there is a 99.9% success rate that a given building block is added, then there will be more than 90% yield of the product.
Achieving a high yield x for each building block addition will likely be a challenging macromolecular design problem. However, it is solvable with sufficient optimization of the building block interfaces. Such optimization has been done among oligonucleotide synthesis companies[30] where per building yields in excess of 99% are routinely achieved. There are also examples of large multi-component materials made from DNA assembled with high per monomer yields[31].
It should be noted that the system would still be useful even if this program does not attain sufficient yield x to make multi-component proteins with one hundred parts. The fundamental macromolecular additive manufacturing process — of anchoring the product to a support — would make it possible to add two or three non-protein modifications to proteins without needing different orthogonal attachment chemistries. In turn, this could unlock a capability that is currently difficult in the biopharmaceutical industry and be used to make advanced antibody drug conjugates with several non-protein materials.
C. Overspecialization of specific building block designs
Development of macromolecular additive manufacturing for a specific protein building block might overspecialize the system to a niche material. It is likely that a substantial amount of time and resources will be needed to optimize the protein building blocks so that they perform well enough to form multi-component proteins that are substantially larger than what is possible with the status quo. This program has to strike a balance optimization of one type of building block versus showing that the system is general enough to use other building blocks and input materials.
This risk must be avoided because it would make the value of the system unclear beyond the immediate research and development in this program. Here are some steps this program is taking to avoid this trap:
- Demonstrate generality of the solid-phase material for handling different macromoles. For instance, the solid-phase support in this program could be further tested to put together multi-component DNA origami structures. Moreover, this program will consider how the composition of the solid-phase supports influences their ability to handle macromolecules with particular physical properties. In turn, it might be possible for this program to deliver actionable rules for selecting support materials for specific macromolecular assembly tasks.
- The design of the protein building blocks will be tailored so that other functionalities can be added to them. For instance, each building block could feature a tag allowing it to grip other protein cargo featuring the complementary tag. Consequently, multi-component proteins made with these building blocks could be likened to molecular ‘breadboards’ much like what has emerged with structural DNA nanotechnology and DNA origami. It might also be possible to design several protein building blocks of differing dimensions, but using the same linkage chemistries, so that multi-component proteins of any desired size could be achieved by mixing and matching building blocks[32].
- The fundamental process difference of this system — where the user can readily exchange liquid reagents around the product during any stage of the reaction — will be applied to add two or more different modifications to the multi-component protein. This is something that is difficult to do with status quo approaches and would provide an alternate use of the system beyond just protein building block assembly. Moreover, the program should think about demonstrating this because of its potential value for biopharmaceutical development.
D. Risks imperiling transition of the system to the biopharmaceutical industry
i. Utility of therapies composed from many components in biopharma is not immediately clear
We speculate that the biopharma industry should care about this program under the premise that this stakeholder wishes to make multi-component proteins of greater complexity. This premise was established by extrapolating trends in the development of protein biologics composed from several distinct parts including: Development of multispecific antibodies, antibody drug conjugate therapies for targeted chemotherapy, and multivalent conjugate vaccines.
However, there is not a consensus of particular therapies that must be developed if 20, 50, or 100 distinct macromolecular components were combined into a single entity[33]. It could be risky to base the future of this program on the capabilities which it could unlock where there is not an agreed ‘killer app.’ Nonetheless, designers in biopharma will only conceive of therapies that the tools available to them are able to manufacture and this is why it is critical to proceed with this program.
Ii. There is no FDA precedent for protein biologics developed entirely using solid-phase supports
There is a high FDA threshold for biopharma companies to meet to get a drug approved and into the market. The FDA dictates that active pharmaceutical ingredients of drugs entering the market have detailed protocols of current good manufacturing practices (cGMP) that the FDA can enforce. It stands to reason that development of completely new drugs with new manufacturing practices would put a higher cost and time burden on companies and disincentivize such approaches. Also, it is notable that there are already examples of using solid-phase synthetic tools to make cGMP peptide therapeutics[34]. Thus, the lack of a cGMP FDA precedent for macromolecular additive manufacturing to make multi-component protein therapeutics is not an insurmountable barrier. If anything, the work which could be done through Speculative Technologies could be used to derisk these processes for a downstream biopharmaceutical company.
VIII. How is the program structured?
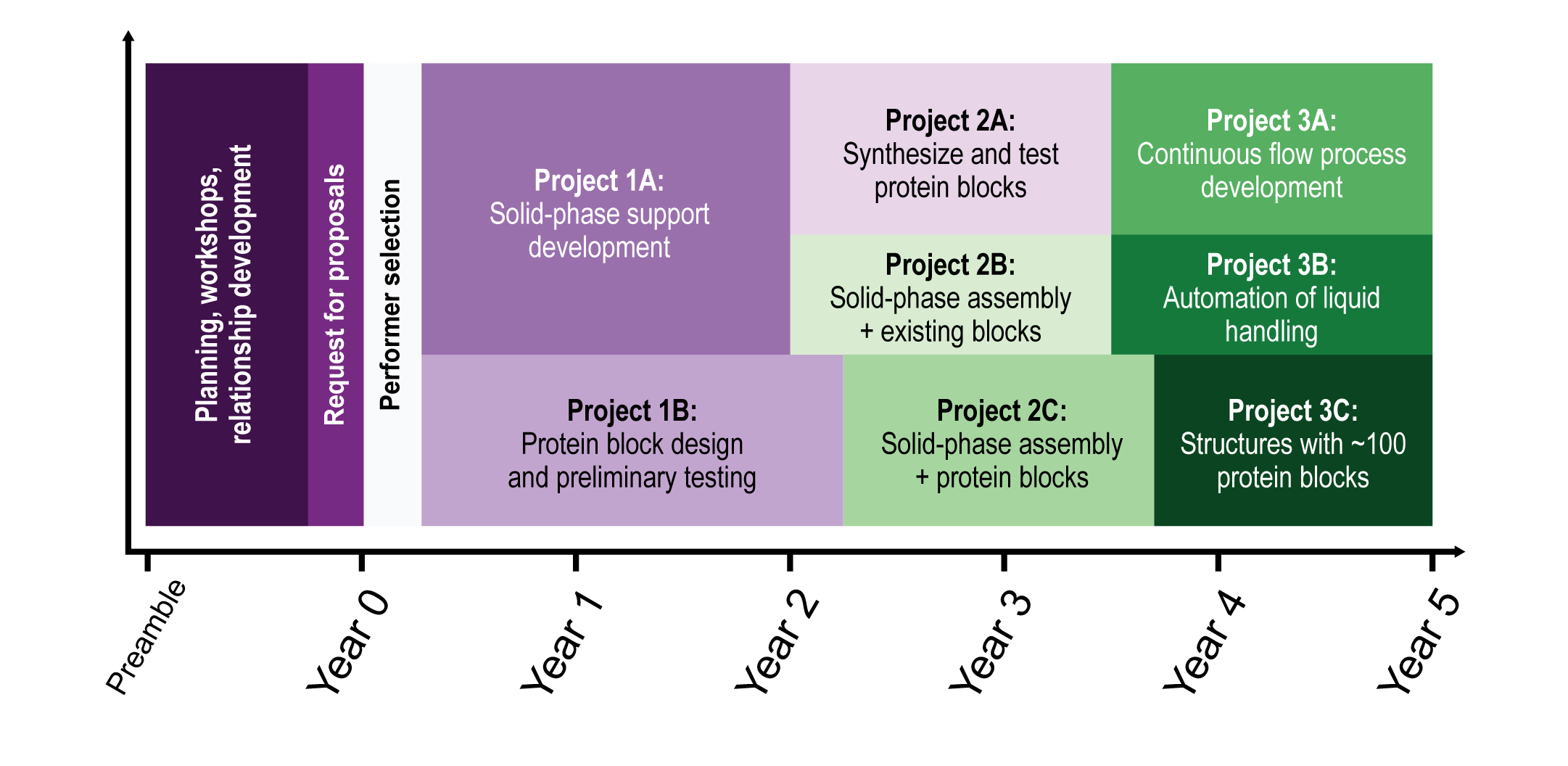
This program will require three phases, approximately five years, and cost about five million dollars, as shown in Figure 6. Each phase is composed of projects, where the product or results from such projects will be necessary for the subsequent phases. The criteria which each of the projects must meet are explained in each phase/project description and as well summarized in the benchmark table.
Selection of the performers that will be doing the work — be it academic researchers, government researchers, startups, or others — will be done in the approach up to phase one. The start times for each phase will be determined by the time to negotiate research agreements, recruit performers that are not already in the Speculative Technologies network, etc. It will also be important that researchers working on a given project be willing to collaborate with researchers handling other projects so as to ensure all components come together in the macromolecular additive manufacturing system.
A. Phase 1: Solid-phase support material development and protein building block development.
Two initial projects over about 1.5 years must be completed to make the materials that will be needed in subsequent phases of the program:
-
A. Solid-phase support material development for macromolecular building blocks. Cost will be about $1.3 million for development of solid-phase support materials.
-
B. Design and preliminary testing of standardized modular protein building blocks. Cost will be about $5ook for a panel of designs and preliminary testing.
i. Project 1A. Solid-phase support material development for large macromolecular building blocks.
The solid-phase supports which currently exist are not suitable for assembling large amounts of multi-component proteins. This project will create new solid-phase supports and is pragmatic about the specific materials used. However, the solid-phase supports must meet the following criteria:
- Loading capacity of ~ 0.1–1 millimole per gram of support material.
- Have sufficiently porosity so that it could be added to a packed bed reactor and allow flow through of reagents, without requiring undue agitation of the solid-phase material.
- Have sufficient phase separation from the bulk liquid solution, such that the solid-phase support can be separated with large pore membranes or standard bench lab techniques such as low speed centrifugation.
- The solid-phase support material must be compatible — that is, not undergo yield limiting adsorption of building blocks or products — with a variety of macromolecules with varied charge properties and secondary structure.
- It is unlikely that a single material would be suitable for every macromolecular building block and it would thus be desirable to be able to tune the properties of solid-phase support on an as-needed basis for the building block in question.
- The material must be stable over a range of temperatures – for instance, -80–90°C — that would be encountered with bio-based macromolecules.
- The solid-phase material must be tolerant of the linkage chemistries selected for the protein building blocks. Consideration should be given on an as-needed basis for resistance to UV light, pH variations, etc.
- It is desirable to covalently attach the first macromolecular building block to the solid-phase support, so as to prevent yield-limiting leaching of the product during reagent handling steps. The strategy must also allow for triggerable release of the product so that it can be recovered after all the blocks have been added.
- The solid-phase material, and method for attaching the product to it, must be able release at least 90% of the product initially anchored to it.
The solid-phase supports from this project are absolutely necessary for the overall success of macromolecular additive manufacturing. Many approaches could be possible including attaching the macromolecules to polymer brushes anchored to a microbead, free polymers that can be triggered to condense from solution during reagent exchange, or large porous resins.
ii. Project 1B. Design and preliminary testing of standardized modular protein building blocks.
This project is needed to create modular protein building blocks to be assembled into multi-component proteins, like plugging toy Lego blocks together. The following design criteria must be considered:
- Each monomeric protein building block must be easy to synthesize and isolate in large quantities. For instance, this could entail using a standard microbial expression (e.g. E. coli) and aim for yields of 10–1000 micromoles per liter of culture.
- The protein building blocks must have the ability to be arranged and connected to form any two dimensional sheet, using whatever lattice structure necessary to make this possible.
- The initial building blocks should be inactive and unable to form higher order structures if mixed together until some inducer is added to trigger their bonding. The intention is to prevent unintentional binding that would lead to yield-limiting misassembly.
- There would need to be a way to covalently link and release one of the macromolecular building blocks to and from the solid-phase support.
- Each building block in a given multi-component protein design must have at least one covalent linkage to another adjacent building block. There must be a contiguous chain of covalent linkages between the building blocks that extend all the way to the solid-phase support.
- A stepwise strategy for adding the blocks one by one on the support into a given design needs to be devised. This requires: (1) Variants of the building blocks that would be needed for particular sites within a design; (2) Complementary bonding interfaces to be designed between the adjoining interfaces of the building blocks; (3) Strategies for activating bonding and triggering covalent linkages between the building blocks. This there are many ways this could be realized, including cleavage of a blocker group from an immobilized building block in a process analogous to solid-phase peptide synthesis, use of a complementary protein “tool” that binds to the interface of adjacent building blocks to contort them and induce a covalent bond, etc.
- Each building block must feature a docking site (orthogonal to the sites for covalent linkage of building blocks to one another) to which other protein cargo can be attached. This could be achieved through either covalent or non-covalent bonds. The intention is to be able to use these multi-component protein arrays as “breadboards” that could be used to position other protein cargo.
This particular project for this stage of the program should be centered on design without fully testing performance of the building blocks. However, there must be a compelling case for why a given design will be able to meet the above criteria for success and experimental demos showing this is strongly encouraged if possible.
B. Phase 2: Implementation of protein building blocks and using the solid-phase supports to make multi-component macromolecules.
Three projects over about 1.5 years will be necessary to demonstrate macromolecular additive manufacturing. The solid-phase support that meets the criteria for this program will be chosen and applied to the following:
-
A. Synthesize protein building blocks, anchor them to a solid-phase support, and develop protocols for linking building blocks to one another. Cost will be about $800k to implement designs of protein building blocks.
-
B. Demonstrate solid-phase assembly of multi-component macromolecules using established building blocks. Cost will be about $500k to apply macromolecular additive manufacturing to building blocks that have been optimized elsewhere.
-
C. Demonstrate solid-phase assembly and modification of multi-component proteins with up to ten different parts. Cost will be about $800k to make multi-component proteins of intermediate complexity.
i. Project 2A. Synthesize protein building blocks, anchor them to a solid-phase support, and develop protocols for linking building blocks to one another.
This project will implement the protein building block designs from project 1B, where the results will be assessed against the criteria for success listed in project 1B. Furthermore:
-
It must be demonstrated that linkages between building blocks can be triggered with high yield. Initially, the performers should be targeting ~90% between each building block, and they should describe a course to boost this to more than 99%. This is necessary so as to make macromolecular additive manufacturing robust enough to create large multi-component proteins.
-
At least one variant of the protein building block must be attached to the solid-phase support from project 1A.
-
A different protein cargo (as described in project 1B) must be attached to the building block and the conjugate building block must be able to assemble with other building blocks.
ii. Project 2B. Demonstrate solid-phase assembly of multi-component macromolecules using established building blocks.
This project will demonstrate that the solid-phase supports from project 1A can be used to assemble large multi-component structures using established building blocks. The rationale is that we need to show the methodology is generalizable and can be used to manufacture materials beyond just the protein building blocks.
Moreover, it is important to develop the processes for macromolecular additive manufacturing so that they may be seamlessly applied to assemble the protein building blocks in project 2C. Working straight with the protein building blocks could be difficult because there will be optimization required to sufficiently increase the linkage yield. By contrast, there are existing macromolecular building blocks made from materials including DNA origami and proteins where the linkage yield has already been optimized for status quo solution-based self assembly.
iii. Project 2C. Demonstrate solid-phase assembly and modification of multi-component proteins with up to ten different parts.
This project will use the materials from project 2A and the methodologies from project 2B to build multi-component proteins that are marginally more complicated that what existing methodologies are capable of. The following would make this project successful:
-
The product structures should contain up to ten distinct building blocks. To our knowledge, the most intricate multi-component proteins made from completely de novo parts with no natural precedent are made from only two different parts, so this achievement would represent a substantial leap in protein complexity.
-
Some of the building blocks should be functionalized with the protein cargo described in project 1B. This demonstration is critical because it highlights the generalizability of the system for building multi-component proteins from different parts — this is important to make the case that macromolecular additive manufacturing could be useful for the biopharmaceutical industry.
C. Phase 3: Continuous flow processes to build and modify large multi-component proteins.
This phase will mature the processes and overall system so that it can build multi-component proteins composed from up to 100 different pieces, which will require three projects over about one and a half years.
- A. Development of continuous flow processes for macromolecular additive manufacturing. Cost will be about $600k to integrate the solid-phase supports into new processes.
- B. Automation of liquid handling where different structures can be made by programming reagent flow. Cost will be about $300k to add automated infrastructure.
- C. Yield optimization of building block addition and use of the system to make multi-component proteins with up to one hundred different parts. Cost will be about $600k to make multi-component proteins with high complexity.
i. Project 3A. Development of continuous flow processes for macromolecular additive manufacturing.
Solid-phase assembly of multi-component proteins requires liquid reagents to be exchanged one or more times each time a building block is added. Hence, this project will devise continuous flow processes to make it possible to create products with dozens or more distinct parts. The challenge in this project will be tailoring the process development to match the properties of the solid-phase material from project 1A. The following properties and specific criteria must be considered:
-
Reagents must be added to and from the system without causing flow-limiting fouling of the solid-phase support. For instance, if the supports were composed of microbeads in a packed-bed reactor, the pressure of fluid flow into the material could damage it to the extent that the product is trapped in the system.
-
It needs to be possible to evenly exchange liquids in the system, such that there are not stagnant areas on the solid-phase support where reagents from a previous step are trapped. Otherwise, there will be a proliferation of incomplete products that will diminish yield.
-
These processes should be demonstrated to be scalable over a 100o-fold range. For example, early demonstrations could entail solid-phase supports occupying ~0.1 milliliters and later demonstrations using ~10 milliliters. It would be important to show linear scaling between the product amount versus reactor chamber volume so as to make a case for the tenability of transitioning the system to industry.
ii. Project 3B. Automation of liquid handling where different structures can be made by programming reagent flow.
The continuous flow process established in project 3A would need to be complemented with automated exchange of liquids and building blocks, so as to make it easy to program formation of different multi-component proteins.
- This goal could be achieved with purpose-built fluid systems that use computer controlled pumps and other infrastructure.
- Alternatively, existing tools from chromatography or other liquid handling tools could be repurposed for the program. However, there would need to be a case for how such approaches could demonstrate scalability of the system.
- Automate the solid support assembly, where beads are situated in workflows that can be handled with programmable liquid handling tools.
iii. Project 3C. Yield optimization of building block addition and use of the system to make multi-component proteins with up to one hundred different parts.
This project will apply the tools and building blocks for the capstone demonstration of this program: To make multi-component protein nanostructures from scratch from about one hundred different parts.
-
This target will require close collaboration between teams working in all the other levels of the program.
-
It will also be important to use the system to make a number of different multi-component proteins, to demonstrate how macromolecular additive manufacturing can readily create different designs by reprogramming the system.
IX. What are the benchmarks for each phase of the program?
The benchmarks listed below are sometimes cross referenced to multiple projects. This indicates projects that might be carried out:
- by the same performer at different points in time,
- by different performers where there might be need to closely collaborate to ensure that the deliverable from one project is a suitable input into the next project,
- or by different performers where each group must meet the same benchmark for their respective project.
We also note that these benchmarks were derived from our best guesses about how to make this program relevant to the broader world and what we think might be possible by applying the research tools before us — there is some room to change these as we learn more!
| Description | Benchmark | Project |
| Loading capacity of product per mass of solid-phase support. | 0.1–1 mmol g-1 | 1A |
| Method of linkage of product to the solid-phase support. | Covalent bond that can be broken on command | 1A/1B/2B |
| Product materials to which the solid-phase support must be compatible with. | Protein building blocks, DNA building blocks | 1A |
| Temperature range over which the solid-phase support must remain stable. | -80–90°C | 1A |
| Other conditions under which solid-phase support might need to remain stable. | Conditions under which building block linkage is triggered (e.g. UV light, pH extremes, etc.) | 1A/1B |
| Fluid pressure under which the solid-phase support would need to remain stable. | To be determined | 1A |
| Geometry of the protein building blocks. | Whatever necessary so they can be arranged into 2D sheets | 1B |
| Number of linkage sites to other protein cargo on each protein building block. | 1 | 1B/2A |
| Yields of protein building blocks per liter of cell culture if manufactured using cellular protein expression. | 10–1000 µmol L-1 | 1B/2A |
| Kinetics of addition for each building block added. | > 105 M-1 s-1 | 1B/2A |
| Yield of addition for each protein building block added to the solid-phase supported assembly. | ~90%, with a viable course to reaching ~99% | 1B/2A/2C/3C |
| Amount of product from solid-phase supports in phase 2 of the program (i.e. push towards max loading capacity of the solid-phase w/ perhaps 1 mg of support in a lab scale volume of ~100 µL). | 1 pmol → 1 µmol | 2B/2C |
| Number of building blocks/modifications in multi-component proteins of intermediate complexity. | ~10 | 2C |
| Scaling behavior of increasing the amount of solid-phase support to make phase 2 intermediate multi-component proteins and scale to attain (i.e. use perhaps 100 mg of support in a larger scale volume of ~10 mL). | Linear, 100 µmol | 3A |
| Amount of product from solid-phase supports in phase 3 of the program (i.e. push towards max loading capacity of the solid-phase w/ perhaps 1 mg of support in a lab scale volume of ~100 µL). | 1 pmol → 1 µmol | 3C |
| Number of building blocks/modifications in multi-component proteins of high complexity. | ~100 | 3C |
X. Conclusions
Macromolecular additive manufacturing might make it possible to build multi-component proteins with ten to one hundred more parts than what can be routinely done with existing approaches. This would be achieved by building the products one building block at a time using solid-phase materials that are specially tailored to handle large macromolecules.
It should also be noted this program is not intended to reject current tools for making proteins. Rather, the program identifies certain multi-component proteins that are impossible to achieve with the status quo tools. It suggests that pragmatic merging of liquid based self-assembly, solid-phase techniques as inspired from synthetic chemistry, protein prediction and design algorithms, and cell based tools to make small proteins offer a viable path to realizing proteins of greater complexity.
In turn, the resulting multi-component proteins could enable artificial ‘antibodies’ that engage many biological targets simultaneously, vaccines made from multiple parts of infectious agents for robust immunity to many pathogens, or rapid prototyping of cell-free factories to synthesize chemicals with spatially organized enzymes.
Speculative technologies cannot bring this to reality alone. We require input from the community to improve this roadmap. Furthermore, we need a talented and committed group of scientists and engineers to fund to do the hard research and development work. Please reach out if you are opinionated about the program or can think of ways to make it happen!
XI. Acknowledgements
The inspiration for macromolecular additive manufacturing is the culmination of ideas from a community of enormously generous people that is too large to name individually. This includes members from industry who helped determine serious contexts of use for the technology and members of the research community who gave insight into how the technology could be built. I am indebted to these folks for their time, insight, and candidness in answering my many questions. You know who you are and thank you!
The specific approaches for making complicated materials from simple building blocks were largely generated by the participants at the January 2023 "Synthetic molecular additive manufacturing" workshop in Austin, TX. In particular, I thank William Shih, Petr Šulc, Andrew Turberfield, Erik Benson, Yonggang Ke, Ariel Ben-Sasson, Kate Ademala, Florian Praetorius, Nicholas Stephanopoulos, Cole DeForest, Alexander Marras, Kyle Meador, and Zhe Li. I also feel indebted to David Baker, Neil King, and the entire University of Washington Institute for Protein Design for their openness in sharing the state of the art of protein design tools. I also thank the people who took the time to read and provide feedback on draft versions of the roadmap. Many of these folks were from the workshop, and I must also give a special shout out to Frances Anastassacos and Jacob Majikes.
Finally, I thank the entire team at Speculative Technologies, whose openness and excitement to bringing sci-fi things into the world is contagious. In particular, I thank Ben Reinhardt for hiring me as the first full time program manager.

Change Log
09-28-2023
- Fixed truncated heading titles in Project 1A and Project 1B subsections.
- Updated the Acknowledgments with a couple missing names.
- Footnote added to explain calculations in the scalability analysis[28:1].
‘Multi-component proteins’ are defined in this roadmap as protein nanostructures composed from two or more different protein parts. In biological contexts, this would be referred to as quaternary protein structure where large complexes are formed from multiple protein subunits or chains. This program also makes the distinction between multi-component proteins that are composed from the multiple copies of the same subunit, versus those that are composed from different subunits. This program is focussed on making multi-component proteins that meet the latter criterion with different subunits — with that, it would become possible to make asymmetrically shaped protein assemblies, which has been an otherwise difficult to achieve objective using existing methods for designing and manufacturing proteins. ↩︎
See Bruce Merrifield’s Nobel Prize winning innovation here. ↩︎ ↩︎
See an overview of peptoids. They were discovered by Ron Zuckermann and colleagues in drug development and there is a community which builds things from peptoids. ↩︎
See an overview of spiroligomers. ↩︎
There is some nuance in the precise use of the term ‘self-assembly.’ There are some people who consider protein folding to be an example of ‘self-organization,’ with the nuance of such terminology described here. Self-assembly (in this roadmap) refers to macromolecules favorably binding to portions of themselves or other macromolecules to assemble into more complicated macromolecular structures. This is mediated by intermolecular forces between the interacting atoms of the various macromolecular pieces, that are in effect encoding formation of a higher order product structure. The finer nuances about ‘self-assembly’ versus ‘self-organization’ are meaningless for this roadmap. ↩︎
The largest single protein made in eukaryotes is the muscular protein called titin, which adds elasticity to muscle and is ~30,000 amino acids in length. By comparison, the hormone insulin is ~50 amino acids long. The abundance of protein versus the length of the RNA transcript coding it are negatively correlated. Such limits of ribosomal protein synthesis might be explained by evolutionary constraints, intrinsic limits of the ribosome itself, and translational regulation. Additionally, it would be challenging for people to create, handle, and integrate large genes into cells for proteins of the scale of titin because of limits in gene synthesis and molecular biology. ↩︎ ↩︎
There is some nuance to consider here. There are many different orthogonal chemistries for attaching things to proteins have been developed and each specific chemistry will require unique reaction conditions that are not necessarily compatible with another linkage chemistry. For instance, one linkage might become cleaved or altered under chemical conditions for another linkage. Moreover, use of such chemistries in the biopharmaceutical industry must be high yielding to support scale up of products and this adds an additional constraint towards esoteric combinations of linkage techniques. ↩︎
The challenge of incorporating structural assembly instructions and other functions should NOT be underestimated. The present algorithms to design parts from proteins are limited even for fitting two different protein subunits together. It is unclear how long it will take for such design algorithms to improve to where it becomes possible to make any new multi-component protein material. ↩︎
Here is an excellent review of where solid-phase techniques have been applied to proteins from Dr. Seah Ling Kuan’s research group. ↩︎
Here is an excellent review of where solid-phase techniques have been applied to proteins. However, the examples within this review seem to be limited to use cases in academic labs. There are no examples to the knowledge of the authors of this roadmap where such techniques have been applied more broadly to products like biopharmaceuticals in the market. ↩︎
Here is an example of where Ralf Seidel’s group used magnetic beads to build up multi-component structures from DNA origami using magnetic beads as a solid-phase support. ↩︎
The US Congressional Budget Office (CBO) prepared a 2021 report describing the research and development trends in the pharmaceutical industry. ↩︎
The Critical Assessment of protein Structure Prediction (CASP) experiments shows this rapid improvement in tools for predicting protein structure from polypeptide sequence. CASP is a community of different research groups who have competed to make better prediction algorithms. ↩︎
Here is one example in structural DNA nanotechnology where researchers build a rendition of the Mona Lisa composed from 64 different DNA origami parts. There was a precipitous dropoff in yield as the total number of parts was increased from four to 16 to 64 parts. ↩︎
The authors in this example built protein squares and triangles from linear protein parts that were purified from cells. Each shape required multiple hierarchical steps where subassemblies were formed from single parts that were subsequently mixed together to build the final shape. ↩︎
The authors here built a printer-like device from DNA origami to selectively expose and activate nodes on a DNA origami sheet to pattern it with other DNA strands. ↩︎
It should be noted that “parts” in this context refers to distinct protein subunits or chains. There are examples of protein therapies composed of multiple protein domains, that might also be described as “parts,” that are concatenated into a single coding sequence where the set of domains are expressed as a single protein. For instance, this work on COBRA T cell engagers combines perhaps seven different domains as a single chain diabody. There is an upper limit to the number of domains which could be combined into a single coding sequence because of the limitations in the length of proteins that cells can manufacture[6:1]. ↩︎
Here is a review which speaks about the challenges and progress towards building spatially organized systems of enzymes. ↩︎
This work describes a simple but compelling model that shows how compartmentalization of a two enzyme system prevents loss of intermediate substrate to increase reaction efficiency. The results are NOT due to proximity of the two enzymes (i.e. direct channeling of the substrate between enzymes), as naive expectations might lead some to believe. ↩︎
Eric Drexler’s Engines of Creation is the foundational text that lays out a vision of nanotechnology where tiny molecular machines assemble macroscale things from the bottom-up atom by atom. ↩︎
The authors here built a device from DNA origami to selectively expose and activate nodes on a DNA origami sheet to pattern it with other DNA strands. A printhead made from DNA origami is rastered along a DNA origami frame where the motion is driven by strand displacement of oligonucleotides. ↩︎
The drug tirzepatide sold under the brand name Mounjaro is lucrative and has been an effective treatment for type II diabetes. This work from Eli Lilly uses a combination of solid-phase peptide synthesis and techniques in free solution to make kilogram amounts of the drug. ↩︎ ↩︎
Brad Pentelute’s group optimized the chemistry and infrastructure for solid-phase peptide synthesis to make polypeptides that were as long as ~170 amino acids. They were able to use this to synthesize functional proteins that were indistinguishable from recombinant protein controls purified from cells. ↩︎ ↩︎
There are solid-phase supports which may be used for long denatured polypeptides. However, these might not be suitable for the larger protein structures which this program is pursuing. ↩︎
Cascade Biocatalysts is developing solid-phase supports that can be tailored to maintain function of enzymes with different biophysical properties. They maintain better activity and lifespan of enzymes versus immobilization strategies preceding theirs. ↩︎
In this work, chemically modified DNA oligonucleotide primers were used to create double stranded DNA segments with polymerase chain reaction (PCR). Subsequently, they used the chemically modified primers to grow polyacrylamide chains that could be condensed on command to recover single strands of DNA. ↩︎
An annotated Google Colab notebook shows how the space time yield estimates were calculated. ↩︎ ↩︎
DNA synthesis companies are motivated to increase yields of their chemistry because it enables them chemically synthesize longer oligonucleotide products that may be of interest to their customers. ↩︎
This work built multi-component DNA origami structures and claimed that origami building blocks could be added with a yield of ~97% per building block added. ↩︎
This idea could be thought of as thinking of the protein building blocks as gauge blocks as used by machinists. ↩︎
One interesting point to consider is that a therapeutic composed from 20, 50, 0r 100 distinct macromolecular parts could have dimensions approaching those of cells and large scale interactions, such as T cell receptor microclusters. This is not attainable with the status quo methods of making protein therapeutics using cells and could represent an interesting context of use for macromolecular additive manufacturing. ↩︎
